Progress
We are generating high quality, annotated assemblies for many. These cultivars were selected by the project principal investigators to represent genetic diversity typical of breeding programs in their agroecological zones.
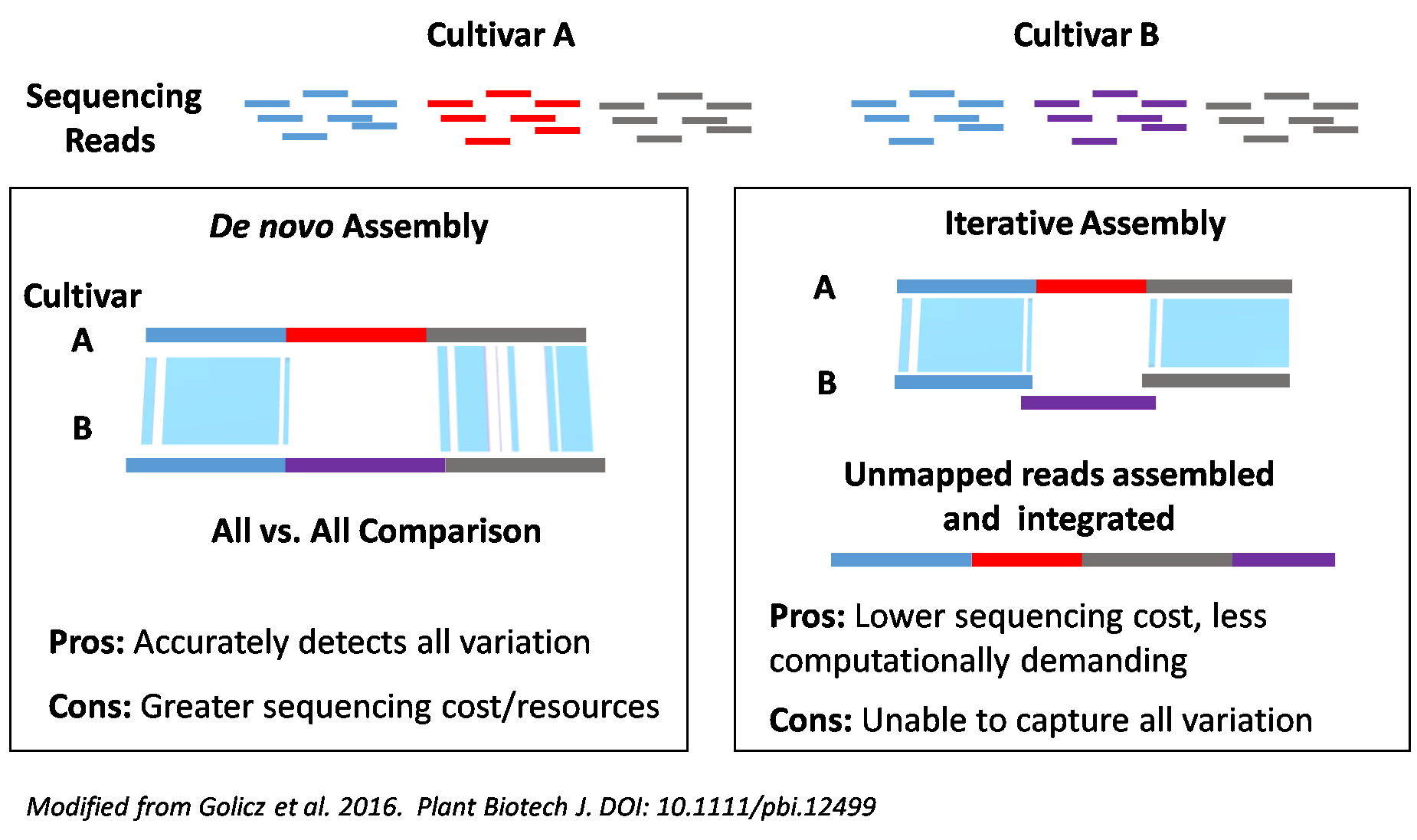
We are using a two pronged approach (see image below). For eight cultivars, we are completing RefSeq assemblies using NRGene’s DeNovoMagicV3.0 assembler. For the remaining cultivars, we are generating assemblies using the open access W2RAP assembly algorithms developed at the Earlham Institute to specifically assemble complex genome from short read sequences . All assemblies generated will be joined into chromosomal sequence using a combination of 10X Genomics data and Hi-C.